MDIPID: Microbiota-drug Interaction and Disease Phenotype Interrelation Database
Accumulating evidence has demonstrated that the human microbiota plays a vital role in disease development and individualized drug response (Nat Rev Microbiol 2024;22:291-308). Indeed, the relationships among microbiota (& its proteins), drugs, and diseases are intricate and bidirectional (Gastroenterology 2023;164:828-40), and an in-depth and thorough understanding of these interactions has increasingly been regarded as a key factor in advancing precision medicine and reducing adverse drug reactions and is attracting considerable attention (Blood 2023;141:2224-38). There are three major types of interactions: (a) Microbiota & its related proteins modulation on drug response (MMDR), which is key for reducing drug-drug interactions and drug resistance (Gut 2020;69:1510-9); (b) Drug or other exogenous substances impact on microbiota (DEIM), which is fundamental to understanding the pathophysiology of disease and improving the efficacy of drug therapy (Cell 2023;186:2705-18 e17); (c) Microbiota-disease associations (MBDA) is essential for identifying predictive biomarkers and helping to stratify patients (Lancet Microbe 2022;3:e969-e83). Since these complex interactions jointly determine drug response and disease development, accumulating relevant information will significantly enhance clinical drug response predictions (Signal Transduct Target Ther 2023;8:201) and provide new insights into disease therapies (Gut 2022;71:1412-25).

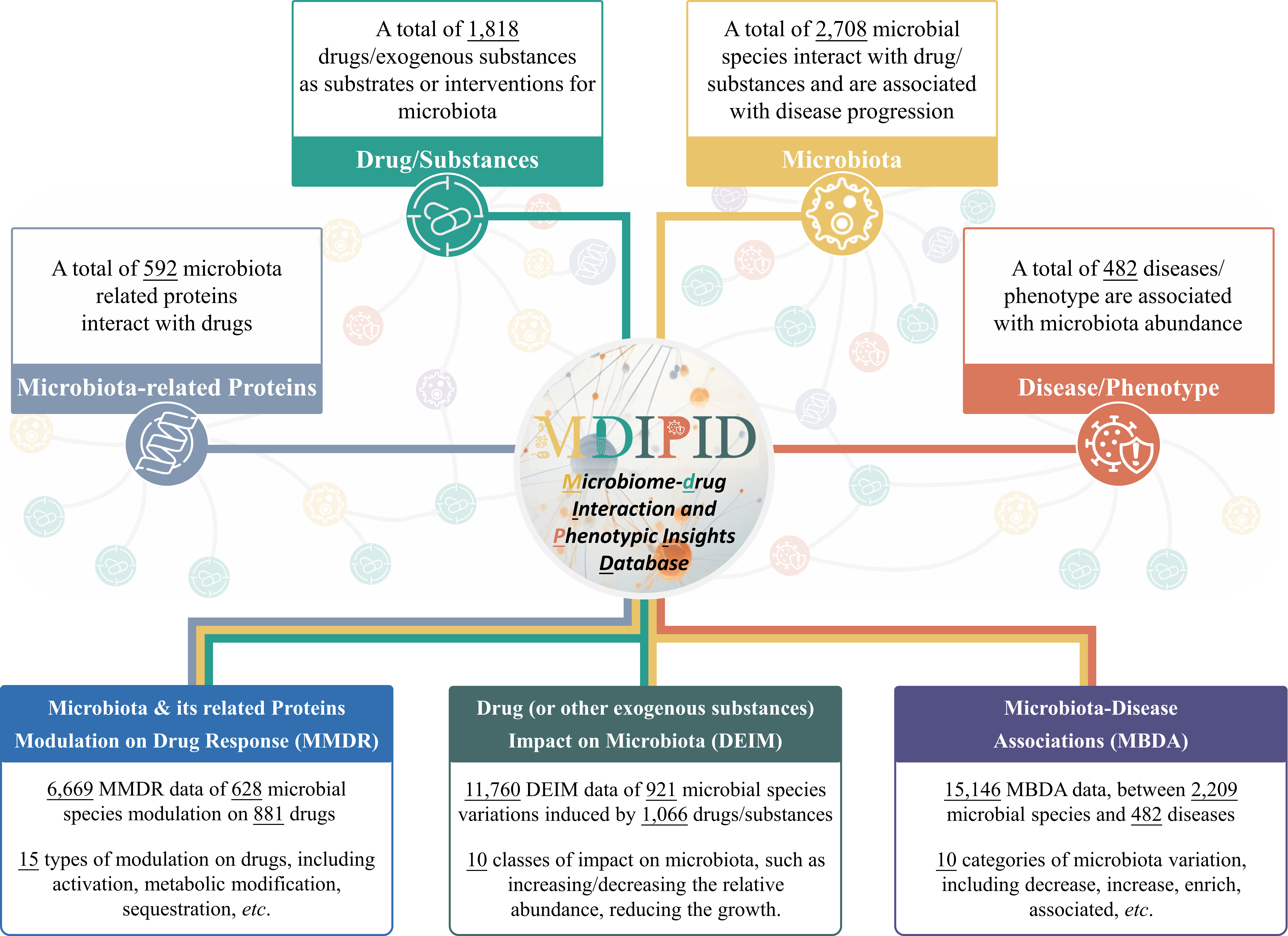
In this study, the Microbiome-drug Interaction and Disease Phenotype Interrelation Database (MDIPID) was therefore introduced to systematically collect and provide the complex bidirectional interactions among microbiota, drugs, and diseases, as well as related proteins & underlying mechanisms (as shown in Figure 1). Particularly, (a) 6,669 MMDR data of 628 microbial species modulation on 881 drugs were described, involving 592 microbiota-related proteins (MRPS) from 282 microbial species and 15 types of drug modulation, including metabolic modification, sequestration and activation; (b) 11,760 DEIM data of 921 microbiota variation induced by 1,066 drugs/exogenous substances were provided, covering 43 groups of substances, such as prebiotics, diet components, and environmental toxicants; (c) 15,146 MBDA data between 2,209 microbial species and 482 diseases were offered, regarding 10 types of microbiota variation, such as decrease, increase, and enrich. Furthermore, the three categories of data that were collected in MDIPID, collectively comprise a comprehensive network, covering 1818 drugs/exogenous substances, 2708 microbial species, 482 diseases, and 592 microbiota-related proteins. All in all, this valuable data described in MDIPID was expected to emerge to be a popular repository that empowers researchers to gain insights into the relationships between drugs, microbiota, and human diseases, ultimately leading to advancements in medicine and the potential for personalized treatment options.